About Our Lab
Our lab studies microbial community ecology ("interactions between microbial populations") and how these contribute to microbial community structure and function, e.g., its resilience towards stresses like pathogen invasion and environmental change. We particularly use symbioses ("different species living together") as model systems where the role of each individual species can be identified and tested, focusing especially on fungus-growing ants. These experiments use both culture and non-culture based microbial ecology techniques (especially genomics) to study microbial community ecology across gradients of environmental and population diversity. We also seek to understand the evolution and ecology of these interactions at the molecular level, focusing especially on the evolution and ecology of natural products and their contribution to community ecology. Besides revealing the molecular underpinnings of community ecology, this molecular research also discovers novel chemicals for drug development.
The community ecology of the T. sepentrionalis symbiosis
 Trachymyrmex sepentrionalis is a fungus-growing ant (tribe Attini) indigenous to the South-Eastern USA ranging from Texas to New Jersey. Fungus-growing ants have an obligate nutritional symbiosis with a specific, co-evolved food fungus (Leucocoprineae) that they host in underground garden chambers, feed with gathered plant material and insect feces, and use as an obligate food source. The garden fungus therefore apparently comprises an "external digestive system" for the ants. Fungus-growing ants also must protect their garden fungus from pathogens, the best studied of which is a second co-evolved fungus Escovopsis. Besides behavioral responses, fungus-growing ants also use symbioses with bacteria that produce antifungal chemicals that inhibit Escovopsis; the best-studied of these bacteria are from the genus Pseudonocardia. Other bacteria symbionts are also likely but less-well studied. Fungus-growing ants therefore host a symbiotic community, representing a simple and experimentally tractable model system and contributing to a novel paradigm for symbiosis highlighting microbiomes instead of simple pair-wise descriptions like "mutualist" and "parasite". This is particularly important to theoretically underpin more complicated symbiosis, e.g., between humans and their microbiota.
Trachymyrmex sepentrionalis is a fungus-growing ant (tribe Attini) indigenous to the South-Eastern USA ranging from Texas to New Jersey. Fungus-growing ants have an obligate nutritional symbiosis with a specific, co-evolved food fungus (Leucocoprineae) that they host in underground garden chambers, feed with gathered plant material and insect feces, and use as an obligate food source. The garden fungus therefore apparently comprises an "external digestive system" for the ants. Fungus-growing ants also must protect their garden fungus from pathogens, the best studied of which is a second co-evolved fungus Escovopsis. Besides behavioral responses, fungus-growing ants also use symbioses with bacteria that produce antifungal chemicals that inhibit Escovopsis; the best-studied of these bacteria are from the genus Pseudonocardia. Other bacteria symbionts are also likely but less-well studied. Fungus-growing ants therefore host a symbiotic community, representing a simple and experimentally tractable model system and contributing to a novel paradigm for symbiosis highlighting microbiomes instead of simple pair-wise descriptions like "mutualist" and "parasite". This is particularly important to theoretically underpin more complicated symbiosis, e.g., between humans and their microbiota.
We take multiple approaches to understand community interactions within the T. sepentrionalis symbiosis. Non-culture based microbial ecology techniques (high-throughput marker gene sequencing and metagenomics) are used to identify in an unbiased manner the microbes associated with T. sepentrionalis and its fungus garden. We pay particularly close attention to differentiating bacterial niches (e.g., particular anatomic structures) and between microbes that are persistently members of the symbiosis vs. those that are transient (i.e., autochthanous vs. allochthanous). These non-culture based experiments guide targeted culturing of each symbiotic member (e.g., cultivar, pathogens, antibiotic-producers, fungus-garden residents). Sequencing sets of targeted genes and (ultimately) whole-genomes from these cultured organisms provides sufficient phylogenetic resolution to understand their population-scale biology and how it impacts symbiotic community structure, complementing less biased non-culture-based techniques without this ability. We also use bioassays between these cultured taxa to create experimental networks of community interactions between these symbionts, the structure of which we correlate to community function. We are particularly interested in how the emergent properties of the entire community govern the evolution of its constituents.
Using symbiosis to understand the ecology and evolution of natural product biosynthesis and resistance, and exploiting it for drug discovery.
Natural products are chemicals that both mediate microbial interactions and are the source and/or inspiration for most commercial drugs. The traditional drug discovery paradigm typically screens collections of microbial culture for novel chemical structures and/or bioactivity. In contrast, we hypothesize that a more fruitful approach is to analyze the chemistry of microbial interactions, e.g., during co-culture of symbiotic partners. We combine such chemical analyses with predictions of the natural product biosynthetic capacity from genome sequences, for which we develop methods that are also widely applicable in other contexts. Finally, we place these natural products within the context of community interactions, identifying where particular molecules evolved to fill specific ecological roles (e.g., to combat a particular pathogen population) and how natural product biosynthetic capacity and resistance co-evolve in the natural environment. This research therefore contributes to drug discovery and to understanding the conditions under which resistance evolves and persists and informs strategies for managing and overcoming this pressing clinical problem.
Wading through the genome deluge
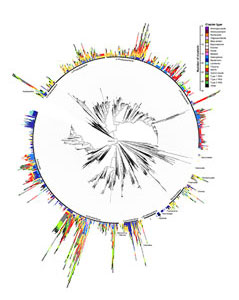 Recent technological advances in DNA sequencing have lowered the cost of genome sequencing to the point where it is becoming the go-to technique for microbial characterization. This has led to the proliferation of these data in public databases that are useful to answer many interesting questions in microbial genome evolution. However, these data are not without their challenges, e.g., due to quality issues resulting from short-read sequencing. We therefore develop software to accommodate these problems, and are particularly interested in problems relating to annotation scalability, amalgamation from multiple data sources, and utilization of draft quality data. This research particularly focus on improving the quality of natural product biosynthetic gene cluster detection and their comparison, thereby facilitating our study of their diversity, evolution, and ecology described above.
Recent technological advances in DNA sequencing have lowered the cost of genome sequencing to the point where it is becoming the go-to technique for microbial characterization. This has led to the proliferation of these data in public databases that are useful to answer many interesting questions in microbial genome evolution. However, these data are not without their challenges, e.g., due to quality issues resulting from short-read sequencing. We therefore develop software to accommodate these problems, and are particularly interested in problems relating to annotation scalability, amalgamation from multiple data sources, and utilization of draft quality data. This research particularly focus on improving the quality of natural product biosynthetic gene cluster detection and their comparison, thereby facilitating our study of their diversity, evolution, and ecology described above.